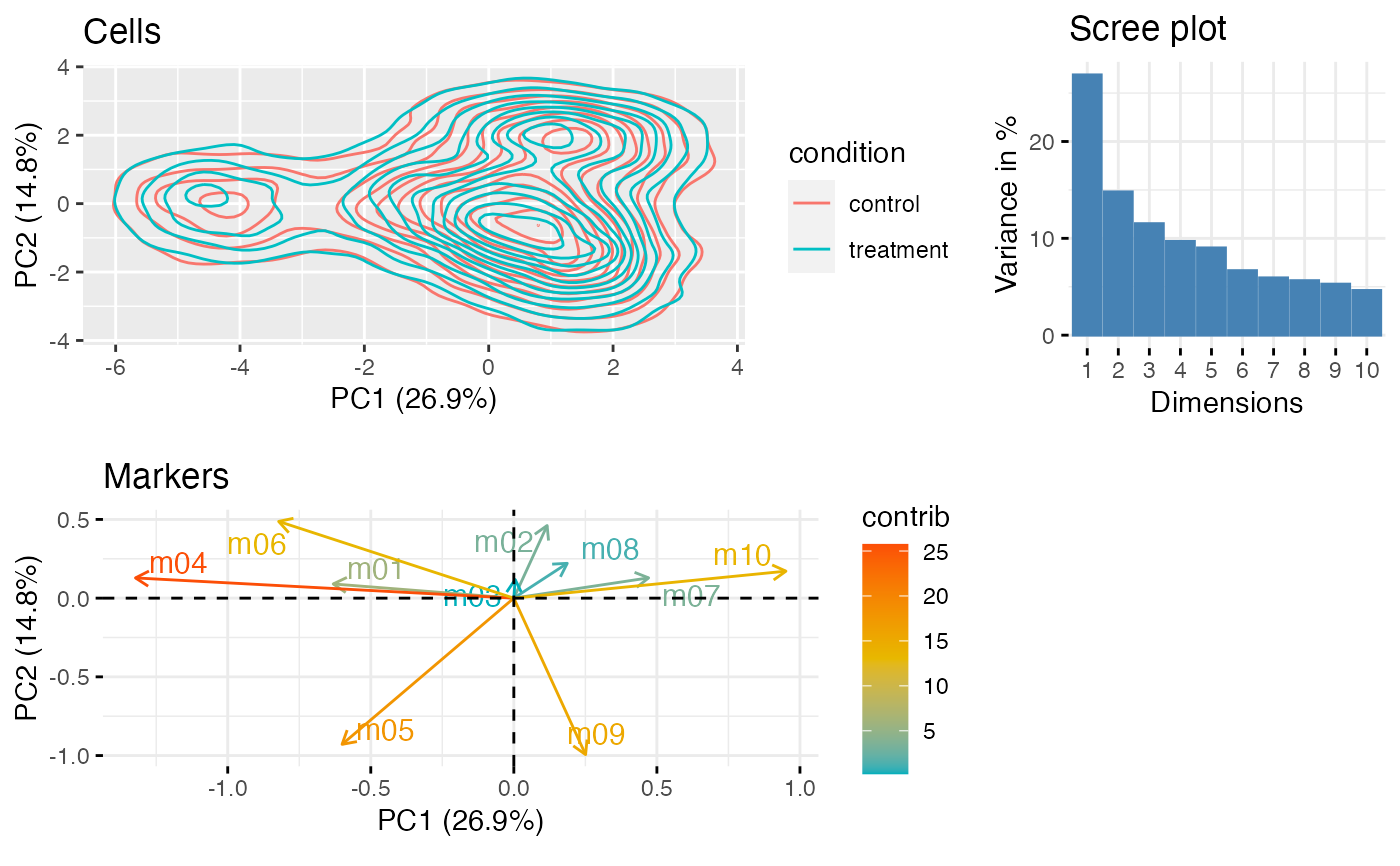
Plot PCA of subsampled data using ggplot
plot_prcomp( df_samples, protein_names, color_var = "treatment", subsample_size = 10000, repel = TRUE )
Arguments
| df_samples | Data frame or tibble with proteins counts, cell condition, and group information |
|---|---|
| protein_names | A vector of column names of protein to use in the analysis |
| color_var | A column name |
| subsample_size | Subsample per color_var variable |
| repel | Repel labels |
Value
cowplot object
Examples
set.seed(23) df <- generate_data() protein_names <- names(df)[3:12] df <- dplyr::mutate_at(df, protein_names, function(x) asinh(x/5)) CytoGLMM::plot_prcomp(df, protein_names = protein_names, color_var = "condition") #> Warning: `group_by_()` was deprecated in dplyr 0.7.0. #> Please use `group_by()` instead. #> See vignette('programming') for more help #> This warning is displayed once every 8 hours. #> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.